This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Gene Ontology
Gene ontology is the study of a gene's existence literally, but practically gene ontology is a standardized terminology either under a biological process, cellular component, or molecular function. This standardization allows for the processes a gene is involved with to be more easily discerned.
Androgen Receptor (AR) Ontology
Using GO consortium, I found that my gene has 14 associations for the human androgen receptor. There are seven biological processes, two cellular components, and five molecular functions involved with AR (see P10275 for associations).
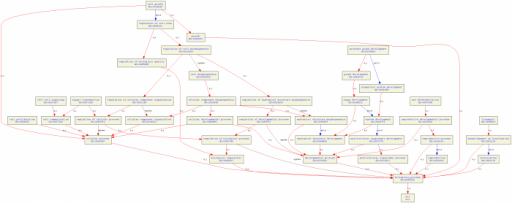
Tree of Biological Processes
Click on the image for a closer look. The schematic shows that AR is involved with cell growth, cell proliferation, cell to cell signaling, prostate gland development, sex differentiation, signal transduction, and transport.
The most expected GO term for the gene is sex differentiation and prostate gland development. AR is involved with both processes (1).
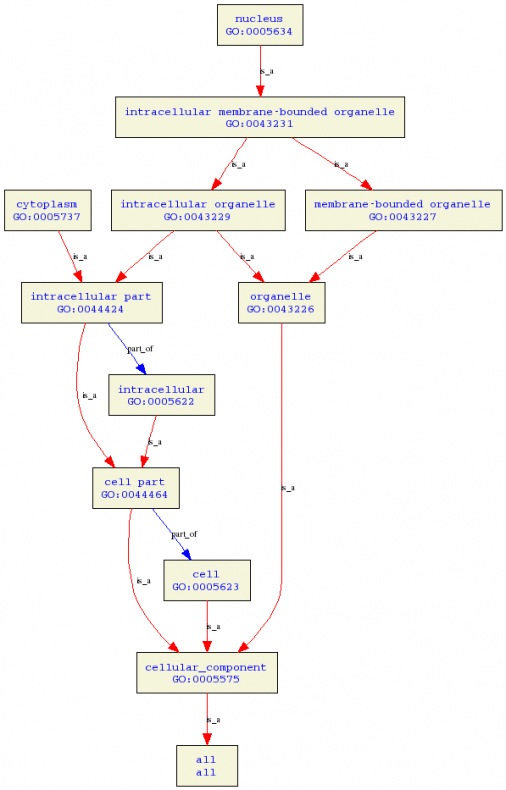
Tree of cellular components
Click on the image for a closer look. AR is located in the cytoplasm and affects the nucleus. AR encounters the androgen in the cytoplasm and then binds to the the nuclear envelope (1).
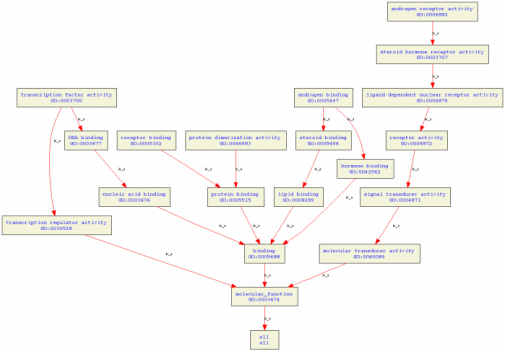
Tree of molecular processes
Click on the image for a closer look. The molecular functions are androgen binding, androgen receptor activity, protein dimerization activity, receptor binding, and transcription factor activity. None of these GO terms are surprising as obviously the androgen receptor is involved with androgen binding and androgen receptor activity (1). The receptor also binds to DNA and acts as a transcription factor, where it becomes a dimer.
References
1. Galani, A., Kitsiou-Tzeli, S., Sofokleous, C., Kanavakis, E., Kalpini-Mavrou, A. (2008). Androgen insensitivity syndrome: clinical features and molecular defects. HORMONES 7(3). Retrieved from: http://hormones.gr/preview.php?c_id=227
Website authored by Sam Trammell. Email: [email protected]. Last updated: February 28, 2009.