This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
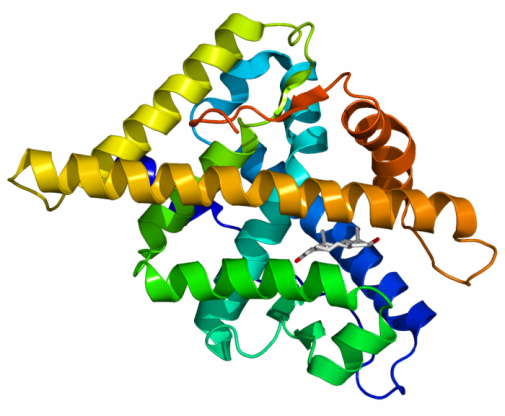
Figure 1: Model of Androgen Receptor
Protein Sequence
The protein sequence used is isoform 1 of the androgen receptor. This protein contains 920 amino acids.
ORIGIN
1 mevqlglgrv yprppsktyr gafqnlfqsv reviqnpgpr hpeaasaapp gasllllqqq
61 qqqqqqqqqq qqqqqqqqqq etsprqqqqq qgedgspqah rrgptgylvl deeqqpsqpq
121 salechperg cvpepgaava askglpqqlp appdeddsaa pstlsllgpt fpglsscsad
181 lkdilseast mqllqqqqqe avsegsssgr areasgapts skdnylggts tisdnakelc
241 kavsvsmglg vealehlspg eqlrgdcmya pllgvppavr ptpcaplaec kgsllddsag
301 kstedtaeys pfkggytkgl egeslgcsgs aaagssgtle lpstlslyks galdeaaayq
361 srdyynfpla lagppppppp phpharikle npldygsawa aaaaqcrygd laslhgagaa
421 gpgsgspsaa assswhtlft aeegqlygpc gggggggggg gggggggggg gggeagavap
481 ygytrppqgl agqesdftap dvwypggmvs rvpypsptcv ksemgpwmds ysgpygdmrl
541 etardhvlpi dyyfppqktc licgdeasgc hygaltcgsc kvffkraaeg kqkylcasrn
601 dctidkfrrk ncpscrlrkc yeagmtlgar klkklgnlkl qeegeasstt spteettqkl
661 tvshiegyec qpiflnvlea iepgvvcagh dnnqpdsfaa llsslnelge rqlvhvvkwa
721 kalpgfrnlh vddqmaviqy swmglmvfam gwrsftnvns rmlyfapdlv fneyrmhksr
781 mysqcvrmrh lsqefgwlqi tpqeflcmka lllfsiipvd glknqkffde lrmnyikeld
841 riiackrknp tscsrrfyql tklldsvqpi arelhqftfd llikshmvsv dfpemmaeii
901 svqvpkilsg kvkpiyfhtq
Physical properties based on primary structure
Using ProtParam, I found the molecular weight to be 99187.8. I found the theoretical pI to be 6, implying that though it is slightly more acidic, it is very close to neutral. The total negatively charged residues is 92, whereas, the total positiviely charged residues is 81. This finding suggests AR is nearly neutrally charged. The estimated half life in humans based on in vitro conditions is 30 hours. I am not sure what the in vivo result would be. However, I believe that it may be shorter in vivo as the protein most likely needs to be tightly regulated as it controls cell division and developmental genes.
Protein Domains
Using SMART, I identified three domains: androgen recep, ZnF_C4, and HOLI. Androgen recep binds to hormones like androgens. The ZnF_C4 domain is the DNA binding domain. HOLI is a lipid biding domain. All of these are consistent with the literature regarding structure and function of the protein (2). The androgen receptor is a cyotplasmic receptor that responds to androgens, triggering nuclear membrane biding and dimerization. This action leads to DNA binding. Altogether, this analysis reveals what is expected.
Using prosite, I identified only the nuclear hormone receptors DND-binding domain. It also predicted with the domain two C4-zinc fingers. This result corraborates with that of SMART. Once again, this result is expected as AR is a nuclear hormone receptor.
Protein modifications
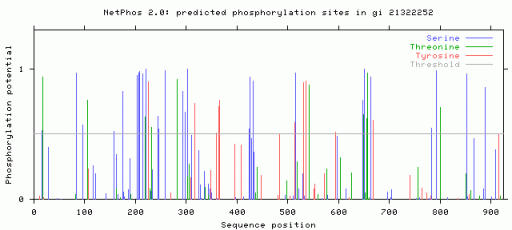
The above figure (generated with NetPhos) shows the likelihood of these sites being phosphorylation sites. As you can see, there are some predicted sites that are unlikely but may actually be phosphorylated in vivo.
Using NetPhos, I identified 27 serine, 10 threonine, and 10 tyrosine phosphorylation sites. This result suggests the involvement of kinases with AR. Most likely upon bindking to dihydrotestosterone, the receptor is phosphorylated so that it may act as a transcription factor. In order to see if AR interacts with kinases, I used NetPhosK. I found several sites interacting with kinases, which is what is expected given the phosphorylation sites shown above. In several locations, several kinases may phosphorylate the site.
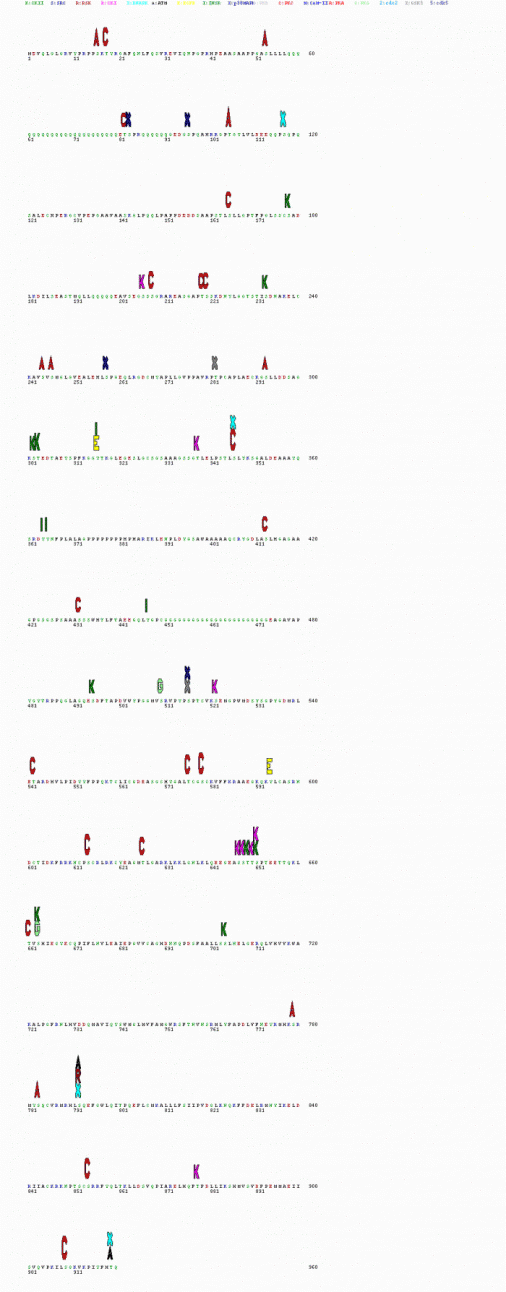
Figure 4: Predicted kinase interactions across protein (generated with NetPhosK)
The following figure shows the predicted kinase interaction sites with the protein. To enlarge, please select the picture. You will also find a key on that page showing what kinases the letters stand for. Notice that the heights of the letters correspond with the prediction values that the site interacts with that kinase.
Localization
Knowing that AR is phosphorylated in the cytoplasm, I wondered how likely the protein is localized in the cytoplasm or other organelles. Using TermiNator, I looked for localization sequences based on the N-terminus. I found a probability of 21.7% that it localizes in the cytoplasm as well as 21.7% that it localizes in the nucleus. This result is expected as it is a nuclear hormone receptor. The other predictions made sense as it is probably modified in these areas. For vesicles of secretory system, the endoplasmic reticulum, and the golgi apparatus, I found a probability of 8.7%. The unlikley prediction that I found was in the mitochondria with a probability of localizing of 13%. This finding seems unlikely as AR is a nuclear hormone receptor.
Protein interactions
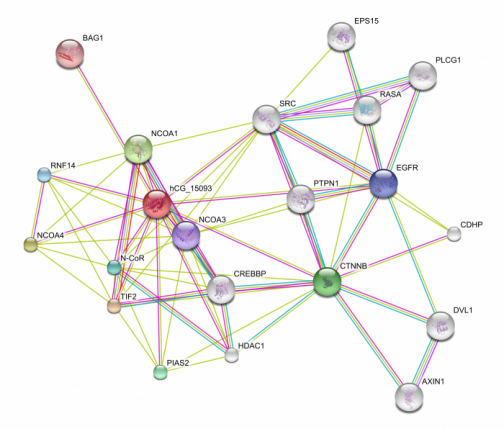
Using String, I found the above protein interactions. The most important immediately important interactions are between the hCG_15093 (androgen receptor) and CTNNB (beta-catenin) and BAG1, EGRFR, DVL1, EPS15, PLCG1, RASA, and SRC. Beta-catenin is involved with the wnt signaling pathway and cell proliferation. BAG-1 is involved with anti-apoptosis. The other interactions listed above are involved with cell proliferation or apoptosis as well. A less obvious but just as important interactions is that between hCG_15093 (AR) and PTPN1. PTPN1 phosphorylates tyrosine residues. This agree entirely with the above findings. Other kinases most likely interact ad can be predicted with String depending on the depth of analysis. These interactions are important for drug targeting. In fact, several anti-cancer drugs do interact with the androgen receptor because of its interactions with these proteins (see phenotype).
Using BioGRID, I found the above interactions as well as BRCA1, also involved with cell proliferation and apoptosis. The interaction between BRCA1 and the androgen receptor with yeast 2 hybridization (Y2H). BAG-1 and CTNNB were found with both Y2H and affinity capture-Western. Because the BRA1 interaction with the androgen receptor was found with Y2H, it may not be a true identification. The other two interactions are likely to be true as they have been confirmed.
Figure 5: Interacting protein newtork (generated with String). The lines indicate the evidence used to discern the interactions. Please select the image to see the key for the evidence lines as well as to enlarge the image and select more detailed information for the proteins.
References
1. (Figure 1) http://androgendb.mcgill.ca/
2. Galani, A., Kitsiou-Tzeli, S., Sofokleous, C., Kanavakis, E., Kalpini-Mavrou, A. (2008). Androgen insensitivity syndrome: clinical features and molecular defects. HORMONES 7(3). Retrieved from: http://hormones.gr/preview.php?c_id=227
Website authored by Sam Trammell. Email: [email protected]. Last updated: April 27, 2009.